Usuario:Renamed user rkdn7i2akc8/prueba imagen doble
Structural DNA nanotechnology[editar]
Structural DNA nanotechnology,[1] sometimes abbreviated as SDN, focuses on synthesizing and characterizing nucleic acid complexes and materials where the assembly has a static, equilibrium endpoint. The nucleic acid double helix has a robust, defined three-dimensional geometry that makes it possible to predict and design the structures of more complicated nucleic acid complexes. Many such structures have been created, including two- and three-dimensional structures, and periodic, aperiodic, and discrete structures.[2][3]
Extended lattices[editar]
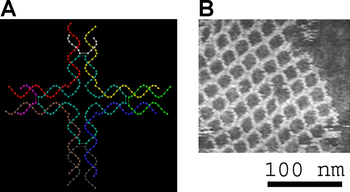
Small nucleic acid complexes can be equipped with sticky ends and combined into larger two-dimensional periodic lattices containing a specific tessellated pattern of the individual molecular tiles.[2] The earliest example of this used double-crossover (DX) complexes as the basic tiles, each containing four sticky ends designed with sequences that caused the DX units to combine into periodic two-dimensional flat sheets that are essentially rigid two-dimensional crystals of DNA.[8][9] Two-dimensional arrays have been made from other motifs as well, including the Holliday junction rhombus lattice,[10] and various DX-based arrays making use of a double-cohesion scheme.[11][12] The top two images at right show examples of tile-based periodic lattices.
Two-dimensional arrays can be made to exhibit aperiodic structures whose assembly implements a specific algorithm, exhibiting one form of DNA computing.[13] The DX tiles can have their sticky end sequences chosen so that they act as Wang tiles, allowing them to perform computation. A DX array whose assembly encodes an XOR operation has been demonstrated; this allows the DNA array to implement a cellular automaton that generates a fractal known as the Sierpinski gasket. The third image at right shows this type of array.[7] Another system has the function of a binary counter, displaying a representation of increasing binary numbers as it grows. These results show that computation can be incorporated into the assembly of DNA arrays.[14]
DX arrays have been made to form hollow nanotubes 4–20 nm in diameter, essentially two-dimensional lattices which curve back upon themselves.[15] These DNA nanotubes are somewhat similar in size and shape to carbon nanotubes, and while they lack the electrical conductance of carbon nanotubes, DNA nanotubes are more easily modified and connected to other structures. [16]One of many schemes for constructing DNA nanotubes uses a lattice of curved DX tiles that curls around itself and closes into a tube.[17] In an alternative method that allows the circumference to be specified in a simple, modular fashion using single-stranded tiles, the rigidity of the tube is an emergent property.[18]
The creation of three-dimensional lattices out of DNA was the earliest goal of DNA nanotechnology, but this proved to be one of the most difficult to realize. Success using a motif based on the concept of tensegrity, a balance between tension and compression forces, was finally reported in 2009.[13][19]
Discrete structures[editar]
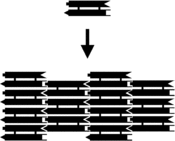
Researchers have synthesized a number of three-dimensional DNA complexes that each have the connectivity of a polyhedron, such as a cube or octahedron, meaning that the DNA duplexes trace the edges of a polyhedron with a DNA junction at each vertex.[20] The earliest demonstrations of DNA polyhedra were very work-intensive, requiring multiple ligations and solid-phase synthesis steps to create catenated polyhedra.[21] Subsequent work yielded polyhedra whose synthesis was much easier. [22]These include a DNA octahedron made from a long single strand designed to fold into the correct conformation,[23] and a tetrahedron that can be produced from four DNA strands in a single step, pictured at the top of this article.[24]
Nanostructures of arbitrary, non-regular shapes are usually made using the DNA origami method. These structures consist of a long, natural virus strand as a "scaffold", which is made to fold into the desired shape by computationally designed short "staple" strands. This method has the advantages of being easy to design, as the base sequence is predetermined by the scaffold strand sequence, and not requiring high strand purity and accurate stoichiometry, as most other DNA nanotechnology methods do. DNA origami was first demonstrated for two-dimensional shapes, such as a smiley face and a coarse map of the Western Hemisphere.[20][25] Solid three-dimensional structures can be made by using parallel DNA helices arranged in a honeycomb pattern,[26] and structures with two-dimensional faces can be made to fold into a hollow overall three-dimensional shape, akin to a cardboard box. These can be programmed to open and reveal or release a molecular cargo in response to a stimulus, making them potentially useful as programmable molecular cages.[27][28]
Templated assembly[editar]
Nucleic acid structures can be made to incorporate molecules other than nucleic acids, sometimes called heteroelements, including proteins, metallic nanoparticles, quantum dots, and fullerenes. This allows the construction of materials and devices with a range of functionalities much greater than is possible with nucleic acids alone. The goal is to use the self-assembly of the nucleic acid structures to template the assembly of the nanoparticles hosted on them, controlling their position and in some cases orientation.[20][29] Many of these schemes use a covalent attachment scheme, using oligonucleotides with amide or thiol functional groups as a chemical handle to bind the heteroelements. This covalent binding scheme has been used to arrange gold nanoparticles on a DX-based array,[30] and to arrange streptavidin protein molecules into specific patterns on a DX array.[31] A non-covalent hosting scheme using Dervan polyamides on a DX array was used to arrange streptavidin proteins in a specific pattern on a DX array.[32] Carbon nanotubes have been hosted on DNA arrays in a pattern allowing the assembly to act as a molecular electronic device, a carbon nanotube field-effect transistor.[33] In addition, there are nucleic acid metallization methods, in which the nucleic acid is replaced by a metal which assumes the general shape of the original nucleic acid structure,[34] and schemes for using nucleic acid nanostructures as lithography masks, transferring their pattern into a solid surface.[35]
- ↑ M. Zadegan, Reza; et, al. (2012). «Structural DNA Nanotechnology: From Design to Applications». Int. J. Mol. Sci. 13 (6): 7149-7162. doi:10.3390/ijms13067149.
- ↑ a b Structural DNA nanotechnology: Seeman, Nadrian C. (November 2007). «An overview of structural DNA nanotechnology». Molecular Biotechnology 37 (3): 246-257. PMC 3479651. PMID 17952671. doi:10.1007/s12033-007-0059-4.
- ↑ Overview: Aldaye, FA; Palmer, AL; Sleiman, HF (26 September 2008). «Assembling materials with DNA as the guide.». Science (New York, N.Y.) 321 (5897): 1795-9. PMID 18818351.
- ↑ Error en la cita: Etiqueta
<ref>no válida; no se ha definido el contenido de las referencias llamadasMao04 - ↑ Other arrays: Strong, Michael (March 2004). «Protein Nanomachines». PLoS Biology 2 (3): e73. PMC 368168. PMID 15024422. doi:10.1371/journal.pbio.0020073.
- ↑ Por favor, pon la referencia que aparece aquí.
- ↑ a b Algorithmic self-assembly: Rothemund, Paul W. K.; Papadakis, Nick; Winfree, Erik (December 2004). «Algorithmic self-assembly of DNA Sierpinski triangles». PLoS Biology 2 (12): 2041-2053. PMC 534809. PMID 15583715. doi:10.1371/journal.pbio.0020424.
- ↑ DX arrays: Winfree, Erik; Liu, Furong; Wenzler, Lisa A.; Seeman, Nadrian C. (6 August 1998). «Design and self-assembly of two-dimensional DNA crystals». Nature 394 (6693): 529-544. Bibcode:1998Natur.394..539W. PMID 9707114. doi:10.1038/28998.
- ↑ DX arrays: Liu, Furong; Sha, Ruojie; Seeman, Nadrian C. (10 February 1999). «Modifying the surface features of two-dimensional DNA crystals». Journal of the American Chemical Society 121 (5): 917-922. doi:10.1021/ja982824a.
- ↑ Other arrays: Mao, Chengde; Sun, Weiqiong; Seeman, Nadrian C. (16 June 1999). «Designed two-dimensional DNA Holliday junction arrays visualized by atomic force microscopy». Journal of the American Chemical Society 121 (23): 5437-5443. doi:10.1021/ja9900398.
- ↑ Other arrays: Constantinou, Pamela E.; Wang, Tong; Kopatsch, Jens; Israel, Lisa B.; Zhang, Xiaoping; Ding, Baoquan; Sherman, William B.; Wang, Xing; Zheng, Jianping; Sha, Ruojie; Seeman, Nadrian C. (21 September 2006). «Double cohesion in structural DNA nanotechnology». Organic and Biomolecular Chemistry 4 (18): 3414-3419. PMC 3491902. PMID 17036134. doi:10.1039/b605212f.
- ↑ Other arrays: Mathieu, Frederick; Liao, Shiping; Kopatsch, Jens; Wang, Tong; Mao, Chengde; Seeman, Nadrian C. (April 2005). «Six-helix bundles designed from DNA». Nano Letters 5 (4): 661-665. Bibcode:2005NanoL...5..661M. PMC 3464188. PMID 15826105. doi:10.1021/nl050084f.
- ↑ a b Error en la cita: Etiqueta
<ref>no válida; no se ha definido el contenido de las referencias llamadasSeemanGrowing - ↑ Algorithmic self-assembly: Barish, Robert D.; Rothemund, Paul W. K.; Winfree, Erik (December 2005). «Two computational primitives for algorithmic self-assembly: copying and counting». Nano Letters 5 (12): 2586-2592. Bibcode:2005NanoL...5.2586B. PMID 16351220. doi:10.1021/nl052038l.
- ↑ Error en la cita: Etiqueta
<ref>no válida; no se ha definido el contenido de las referencias llamadasFeldkamp - ↑ DNA Nanotubes: Lo, PK; Karam, P; Aldaye, FA; McLaughlin, CK; Hamblin, GD; Cosa, G; Sleiman, HF (April 2010). «Loading and selective release of cargo in DNA nanotubes with longitudinal variation.». Nature chemistry 2 (4): 319-28. PMID 21124515.
- ↑ DNA nanotubes: Rothemund, Paul W. K.; Ekani-Nkodo, Axel; Papadakis, Nick; Kumar, Ashish; Fygenson, Deborah Kuchnir & Winfree, Erik (22 December 2004). «Design and Characterization of Programmable DNA Nanotubes». Journal of the American Chemical Society 126 (50): 16344-16352. PMID 15600335. doi:10.1021/ja044319l.
- ↑ DNA nanotubes: Yin, P.; Hariadi, R. F.; Sahu, S.; Choi, H. M. T.; Park, S. H.; Labean, T. H.; Reif, J. H. (8 August 2008). «Programming DNA Tube Circumferences». Science 321 (5890): 824-826. Bibcode:2008Sci...321..824Y. PMID 18687961. doi:10.1126/science.1157312.
- ↑ Three-dimensional arrays: Zheng, Jianping; Birktoft, Jens J.; Chen, Yi; Wang, Tong; Sha, Ruojie; Constantinou, Pamela E.; Ginell, Stephan L.; Mao, Chengde et al. (3 September 2009). «From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal». Nature 461 (7260): 74-77. Bibcode:2009Natur.461...74Z. PMC 2764300. PMID 19727196. doi:10.1038/nature08274.
- ↑ a b c Overview: Por favor, pon la referencia que aparece aquí.
- ↑ DNA polyhedra: Zhang, Yuwen; Seeman, Nadrian C. (1 March 1994). «Construction of a DNA-truncated octahedron». Journal of the American Chemical Society 116 (5): 1661-1669. doi:10.1021/ja00084a006.
- ↑ DNA Boxes: Aldaye, FA; Sleiman, HF (7 November 2007). «Modular access to structurally switchable 3D discrete DNA assemblies.». Journal of the American Chemical Society 129 (44): 13376-7. PMID 17939666.
- ↑ DNA polyhedra: Shih, William M.; Quispe, Joel D.; Joyce, Gerald F. (12 February 2004). «A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron». Nature 427 (6975): 618-621. Bibcode:2004Natur.427..618S. PMID 14961116. doi:10.1038/nature02307.
- ↑ DNA polyhedra: Goodman, Russel P.; Schaap, Iwan A. T.; Tardin, C. F.; Erben, Christof M.; Berry, Richard M.; Schmidt, C.F.; Turberfield, Andrew J. (9 December 2005). «Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication». Science 310 (5754): 1661-1665. Bibcode:2005Sci...310.1661G. PMID 16339440. doi:10.1126/science.1120367.
- ↑ Error en la cita: Etiqueta
<ref>no válida; no se ha definido el contenido de las referencias llamadasrothemundorigami - ↑ DNA origami: Douglas, Shawn M.; Dietz, Hendrik; Liedl, Tim; Högberg, Björn; Graf, Franziska; Shih, William M. (21 de mayo de 2009). «Self-assembly of DNA into nanoscale three-dimensional shapes». Nature 459 (7245): 414-418. Bibcode:2009Natur.459..414D. PMC 2688462. PMID 19458720. doi:10.1038/nature08016.
- ↑ DNA boxes: Andersen, Ebbe S.; Dong, Mingdong; Nielsen, Morten M.; Jahn, Kasper; Subramani, Ramesh; Mamdouh, Wael; Golas, Monika M.; Sander, Bjoern et al. (7 de mayo de 2009). «Self-assembly of a nanoscale DNA box with a controllable lid». Nature 459 (7243): 73-76. Bibcode:2009Natur.459...73A. PMID 19424153. doi:10.1038/nature07971.
- ↑ DNA boxes: Ke, Yonggang; Sharma, Jaswinder; Liu, Minghui; Jahn, Kasper; Liu, Yan; Yan, Hao (10 June 2009). «Scaffolded DNA origami of a DNA tetrahedron molecular container». Nano Letters 9 (6): 2445-2447. Bibcode:2009NanoL...9.2445K. PMID 19419184. doi:10.1021/nl901165f.
- ↑ Overview: Por favor, pon la referencia que aparece aquí.
- ↑ Nanoarchitecture: Zheng, Jiwen; Constantinou, Pamela E.; Micheel, Christine; Alivisatos, A. Paul; Kiehl, Richard A.; Seeman Nadrian C. (July 2006). «2D Nanoparticle Arrays Show the Organizational Power of Robust DNA Motifs». Nano Letters 6 (7): 1502-1504. Bibcode:2006NanoL...6.1502Z. PMC 3465979. PMID 16834438. doi:10.1021/nl060994c.
- ↑ Nanoarchitecture: Park, Sung Ha; Pistol, Constantin; Ahn, Sang Jung; Reif, John H.; Lebeck, Alvin R.; Dwyer, Chris; LaBean, Thomas H. (October 2006). «Finite-size, fully addressable DNA tile lattices formed by hierarchical assembly procedures». Angewandte Chemie 118 (40): 749-753. doi:10.1002/ange.200690141.
- ↑ Nanoarchitecture: Cohen, Justin D.; Sadowski, John P.; Dervan, Peter B. (22 October 2007). «Addressing single molecules on DNA nanostructures». Angewandte Chemie International Edition 46 (42): 7956-7959. PMID 17763481. doi:10.1002/anie.200702767.
- ↑ Nanoarchitecture: Maune, Hareem T.; Han, Si-Ping; Barish, Robert D.; Bockrath, Marc; Goddard III, William A.; Rothemund, Paul W. K.; Winfree, Erik (January 2009). «Self-assembly of carbon nanotubes into two-dimensional geometries using DNA origami templates». Nature Nanotechnology 5 (1): 61-66. Bibcode:2010NatNa...5...61M. PMID 19898497. doi:10.1038/nnano.2009.311.
- ↑ Nanoarchitecture: Por favor, pon la referencia que aparece aquí.
- ↑ Nanoarchitecture: Por favor, pon la referencia que aparece aquí.